Projects
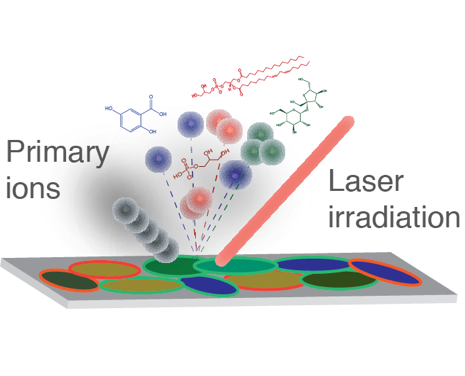
Beam Ionization: Who Gets the Charge and Why Do Some Get Broken Up About It?
Science Objectives
- Elucidate the role of cationization in beam-based desorption/ionization of molecules from surfaces.
- Reveal how the molecular structure regulates its ability to be desorbed and ionized from a surface via beam-based probes.
- Determine how mixtures of molecules modulate desorption and ionization properties from surfaces
Publications:
-
Hoshin Kim, Brittney L. Gorman, Michael J. Taylor, Christopher R. Anderton; Atomistic simulations for investigation of substrate and salt effects on lipid in-source fragmentation in secondary ion mass spectrometry: A follow-up study. Biointerphases 1 January 2024; 19 (1): 011003. https://doi.org/10.1116/6.0003281
Development of Computational Software for High Precision Collisional Cross Section Measurements
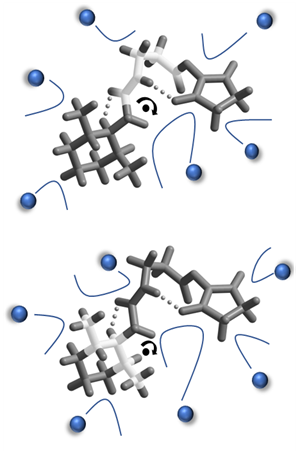
Science Objectives
- Ultra-high-resolution ion mobility experimentally demonstrated the ability to resolve between ions that differ in mobility by only a few ppm.
- Mobility differences can be attributed to differences in rotational properties which are not accounted for in current computational approaches, all of which fail to reproduce experimental results.
- Develop efficient computational approach to calculate collisional cross sections while accounting for the rotational properties of the target ions.
- The mobility dependence on the ion’s center of mass and moment of inertia will be characterized and possibly exploited to gain insights on 3-D structure.
Publications
- Harrilal C.P. V.D. Gandhi G. Nagy X. Chen M.G. Buchanan R. Wojcik and C.R. Conant et al. 2021. Measurement and Theory of Gas Phase Ion Mobility Shifts Resulting from Isotopomer Mass Distribution Changes." Analytical Chemistry. 93, no. 45:14966–14975. doi:10.1021/acs.analchem.1c01736
Ion Confinement at Atmospheric Pressure Using Nonlinear Direct Current Electric Fields
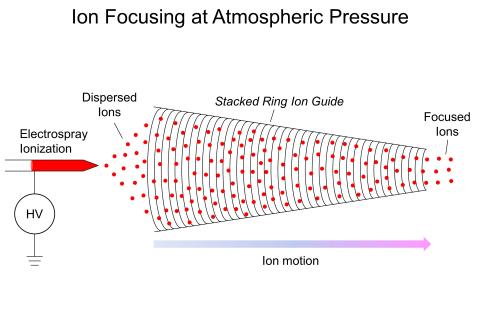
PI: Adam Hollerbach
Science Objectives
Develop method to focus and confine ions at atmospheric pressure (760 Torr) without the use of high voltage radiofrequencies.
- Reduces the need for bulky and expensive vacuum systems to perform ion analysis
- Enables ion mobility studies to be performed at higher pressures without ion losses, allowing for higher ion-mobility spectrometry resolution to be obtained than at lower pressures
- Establishes fundamental understanding of ion confinement effects at elevated pressures
Robust Data Analysis Tool for Locating Lipid Carbon-Carbon Double Bonds and Isomer Separation
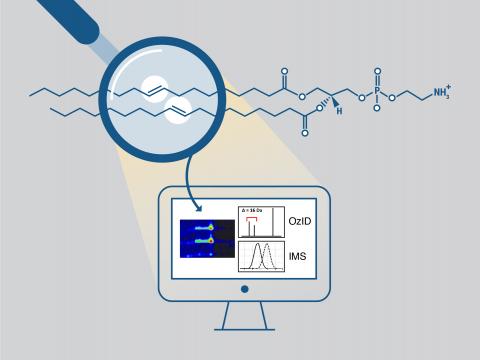
PI: Xueyun Zheng
Science Objectives
Develop robust bioinformatics tools that allow fast data processing and analysis of lipidomics data for confident identification and complete structure elucidation of lipids in complex samples.
- Develop new informatic tool for automated elucidation of double bond locations in unsaturated lipids from ozone-induced dissociation mass spectrometry data.
- Develop a combined DDA-DIA lipidomics data acquisition and analysis workflow using ion mobility spectrometry-mass spectrometry.
Publications:
- MZA: A Data Conversion Tool to Facilitate Software Development and Artificial Intelligence Research in Multidimensional Mass Spectrometry”, Journal of Proteomics Research, 2023, 22, 2, 508–513, https://doi.org/10.1021/acs.jproteome.2c00313
- “mzapy: An Open-Source Python Library Enabling Efficient Extraction and Processing of Ion Mobility Spectrometry-Mass Spectrometry Data in the MZA File Format”, Analytical Chemistry, 2023, 95, 25, 9428–9431, https://doi.org/10.1021/acs.analchem.3c01653
- “LipidOz Enables Automated Elucidation of Lipid Carbon–Carbon Double Bond Positions from Ozone-Induced Dissociation Mass Spectrometry Data”, Communications Chemistry, 2023, 6, 74, https://doi.org/10.1038/s42004-023-00867-9
- “Evaluating Software Tools for Lipid Identification from Ion Mobility Spectrometry–Mass Spectrometry Lipidomics Data”, Molecules, 2023, 28(8), 3483, https://doi.org/10.3390/molecules28083483
Ion Manipulation at Atmospheric Pressure for Increased Reaction Time and Improved Sensitivity
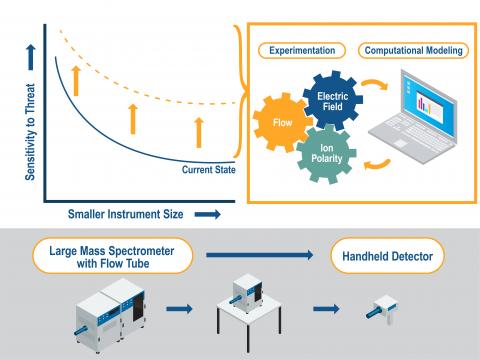
PI: Elizabeth Denis
Science Objectives
Improve sensitivity for mass spectrometry and ion mobility spectrometry by manipulating ions at atmospheric pressure.
- Increase ionization times and ion transfer efficiencies by adjusting gas flows and electric fields at atmospheric pressure.
- Increase ion density by co-mingling oppositely charged ions.
- Compare experimental results with ion trajectory modeling and molecular modeling to advance the fundamental understanding of ion motion at atmospheric pressure.
Publications:
- Ewing R.G., G.L. Hart, M.K. Nims, S.E. Murphy, S. Johnson, J. Chun, and E.H. Denis. 2023. "Reducing ion diffusion at atmospheric pressure through intermingled positive and negative ions." International Journal of Mass Spectrometry 492, 117115. https://doi.org/10.1016/j.ijms.2023.117115
Identification of Molecular Samples via Relational Hypergraph and Topological Models of Multi-Dimensional Mass Spectral Data
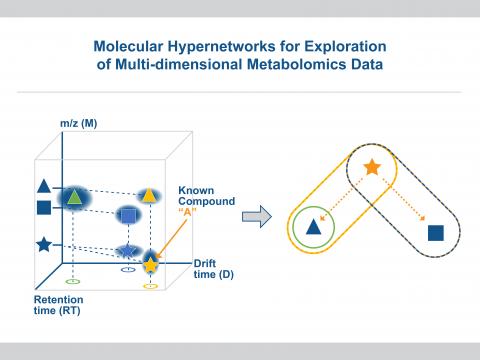
PI: Cliff Joslyn
Science Objectives
Use hypergraph and topological data structures to model and compare multi-dimensional mass spectrometry (e.g., liquid chromatography–ion-mobility spectrometry–tandem mass spectrometry (LC-IMS-MS/MS)) data sets to support sample identification and library matching for improved characterization of unknown samples, reflecting true data complexity.
- Perform subject-matter expert-guided exploratory analysis of the hypergraph structure of LC-IMS-MS/MS data tensors.
- Perform SME-guided exploratory analysis of the topological structure of LC-IMS-MS/MS data tensors, modeled as a multidimensional point cloud.
- Develop initial methodology for the use of hypergraph and topological models to compare LC-IMS-MS/MS data from an unknown sample to a library of similar data from known base samples.
Domain-Aware Normalization and Batch Effect Quantification and Correction for Small Molecules
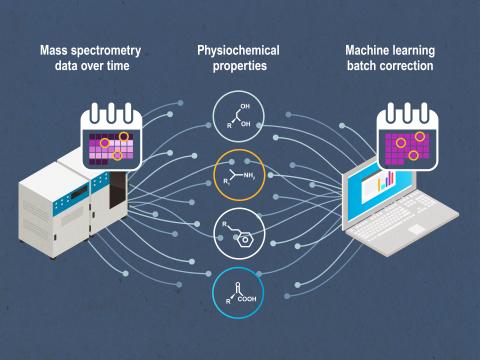
PI: Lisa Bramer
Science Objectives
- Improve evaluate normalization methods for quantified metabolomics and lipidomics datasets.
- Developing effective batch correction methods by incorporating small molecule physiochemical properties, thus enhancing the value of experimental data and enable cutting-edge computational methods such as artificial intelligence.
Publications
Leach, D. T., Stratton, K. G., Irvahn, J., Richardson, R., Webb-Robertson, B. J. M., & Bramer, L. M. (2023). malbacR: A Package for Standardized Implementation of Batch Correction Methods for Omics Data. Analytical Chemistry https://doi.org/10.1021/acs.analchem.3c01289
Enabling Standards-Free, Sensitive, and Accurate Identification of Small Molecules by Ultra-High Resolution Ion Mobility and Highly Accurate Mass Measurements
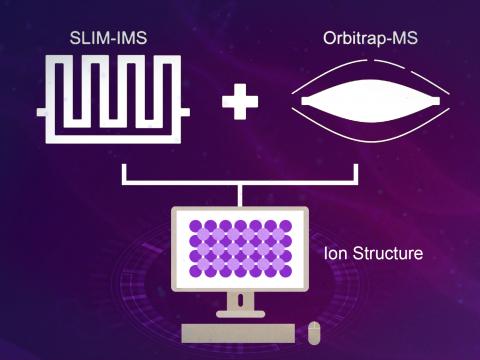
PI: Adam Hollerbach
Science Objectives
Merge traveling wave ion mobility spectrometry employing structures for lossless ion manipulations (SLIM-IMS) with orbitrap mass spectrometry (Orbitrap-MS) to transform our ability to predict the molecular properties of unknown molecules.
- Enable ultrahigh resolution ion mobility separations and high-resolution mass analysis in a single instrument
- Utilize high resolution data sets to provide feedback to enhance predictive models that utilize collision cross section and mass information.
Heracles: In Silico Predictive Tools for Opioid Crisis Intervention
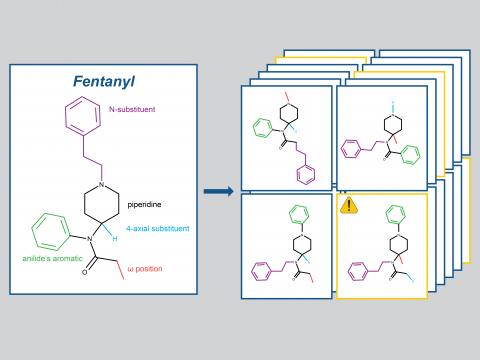
PI: Kate Schultz
Science Objectives
- Build a database of potential fentanyl analogs using chemical combinatorics approaches.
- Develop a computational risk assessment pipeline to determine the relative danger of potential fentanyl analogs .
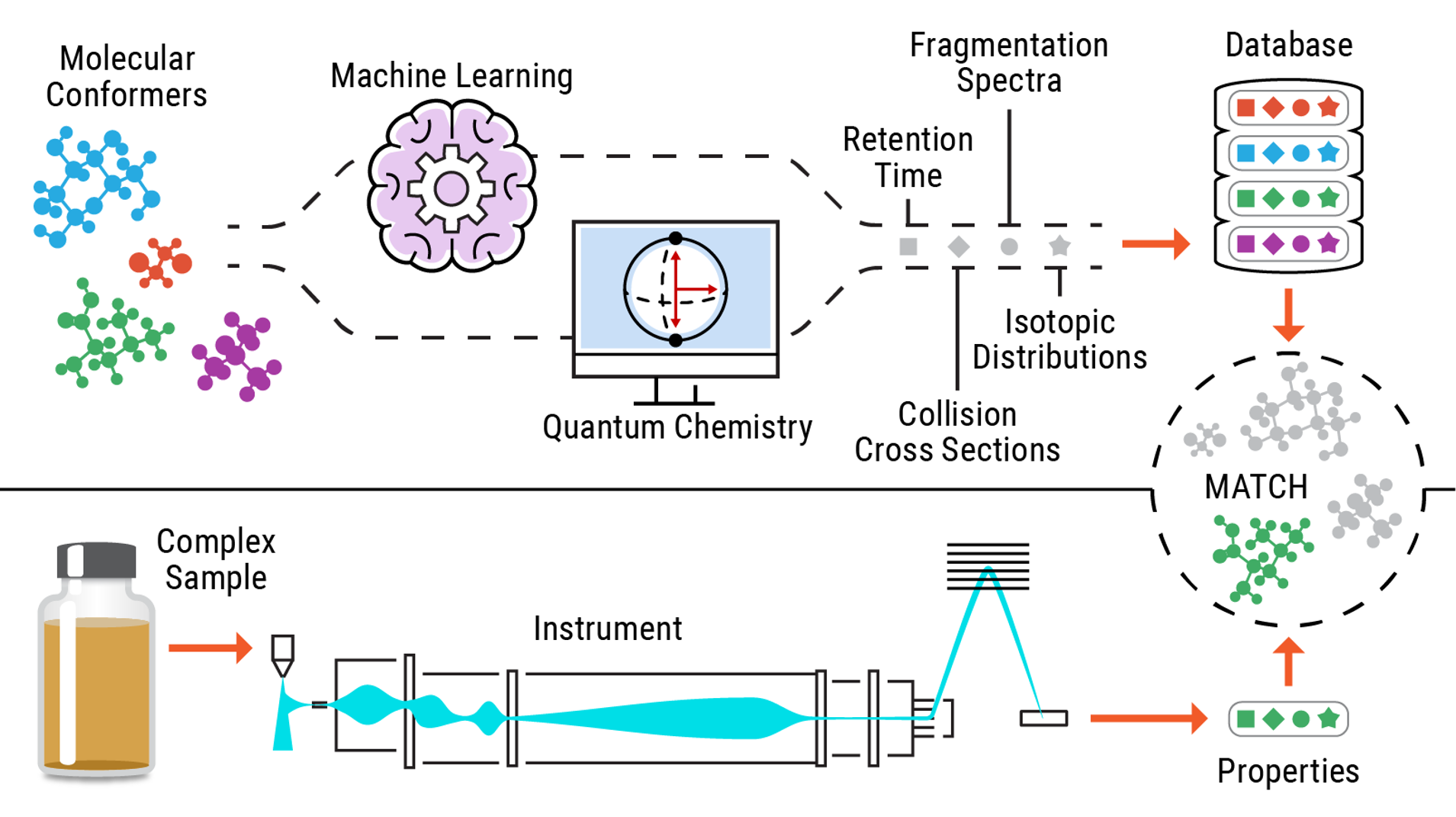
Multi-Dimensional Molecular Identification and Prediction Using IR-IMS-MS and Quantum Chemistry and Machine Learning Approaches
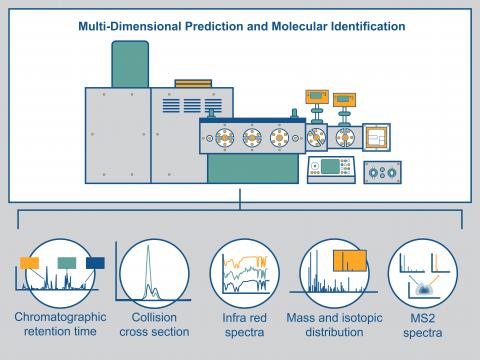
PI: Chris Harrilal
Science Objectives
Unambiguous identification of unknown compounds without authentic reference materials requires multi-dimensional measurements of molecular properties. We are utilizing multi-dimensional information such as the collision cross-section, m/z, MS/MS, and infrared spectra in conjunction with quantum chemical approaches for confident molecular identification.
- Integrate ultra-high resolution ion mobility separations with gas-phase cryogenic infrared spectroscopy and mass spectrometry.
- Develop a computational approach to predict the identity of unknown small molecules based on the multi-dimensional molecular measurements of ions.
Computational Methods for Modeling Electrospray Microdroplet Chemistry for Improved Quantitative Mass Spectrometry
PI: Samantha Johnson
Science Objectives
- Track the evolution of droplet concentration of analytes from bulk concentration inside an electrospray ionization (ESI) capillary.
- Track evolution of analytes in microdroplets to predict spectra.
- Bridge the scales by correlating bulk concentrations, ESI droplets and predicted spectra.
Statistically-Driven Experimental Design to Improve Reference-Free Quantitation of Small Molecules by Liquid Chromatography-Mass Spectrometry
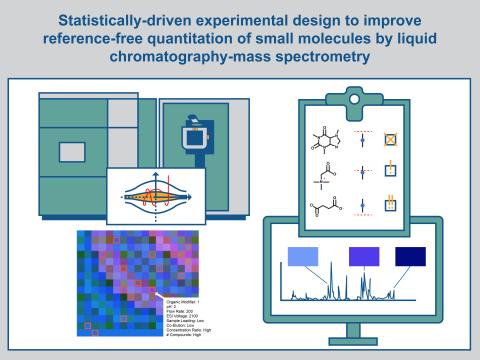
PIs: Fanny Chu and Jessica Bade
Science Objectives
- Improve unknown concentration estimations from mass spectra using machine learning
- Create a comprehensive LC-MS/MS dataset across instrument and sampling parameters to characterize influence on concentration estimations
- Reduce the uncertainty bounds in concentration estimation by improving our understanding of the effects of instrument parameters on LC-MS/MS data
Instrumentation Independent Spectral Prediction with Quantum Chemistry Informed Machine Learning
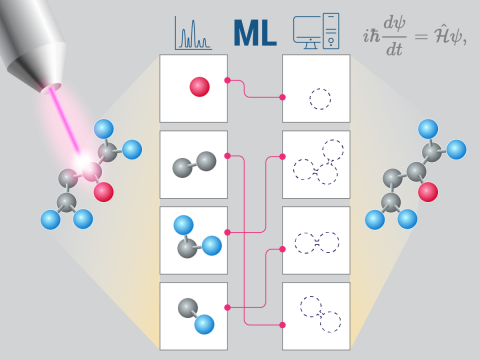
Science Objectives
- Predict the evolution of mass spectrometry peaks across different collision energies for different instrument types.
- Identify the machine readable representations of chemical information that lead to the strongest improvements in machine learning predictions of spectra.
- Develop a pipeline that combines quantum chemistry, machine learning, and experimental data to generate highly accurate reference free spectra.
Computationally-driven discovery and identification of small molecules in biological systems
PI: Jamie Nunez
Science Objectives
-
Develop software to streamline suspect library creation for specific organisms (minimize unknown knowns)
-
Produce initial deep learning network that can be adapted to generate structures from measurement data
Development of a Robust Score and False Discovery Rate for Metabolite Identification
PI: Chaevien Clendinen
Science Objectives
-
Correct conclusions from measurements
-
Use machine learning-based integration of spectral and chemical properties to improve molecular identification confidence with an estimation of overall FDR
- Increase confidence in molecular identifications and significantly reduces the dependency on the analyst, thereby allowing for full automation of data processing steps
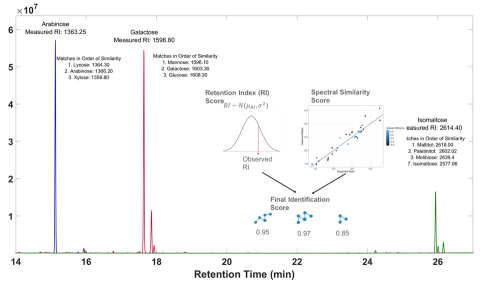
Characterizing identification probability and precision in reference-free compound annotation for metabolomics
PI: Dylan Ross
Science Objectives
•Comprehensive database of molecular properties (m/z, RT, CCS, MS/MS) curated from literature
•High-performance computational pipeline for predicting molecular properties
•Identification probability analysis for probing impact of measurement precision
•Quantitative understanding of relationship between measurement precision and identification probability in reference-free compound annotation
Multimodal, multitask retrieval of molecular structure from measured signatures for reference-free compound identification
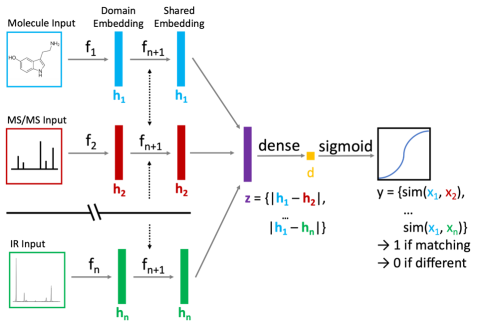
PI: Sean Colby
Science Objectives
- Deep learning-based molecule:signature recognition; utilizes one or more of NMR, IR, UV, MS/MS, CCS to motivate probabilistic identification.
- Rapid triage of known and unknown threats directly from one or more measured signatures; removes reference-based identification requirement.
- Strengthen understanding of molecule:signature relationship; motivate selection/prioritization of critical measurement technology.
Reaction Roulette: Utilizing Elemental MS/MS for Fundamental Characterization of Gas Phase Ion- Molecule Interactions

PI: Khadouja Harouaka
Science Objectives
-
Established library of ion reactivity across the periodic table with several gases and tied the reactivity with instrument tunable parameters
-
Greatly reduced chemical purification, faster sample measurement and overall higher analysis throughput
-
Development of predictive, machine learning capability for analyte-specific measurements
Publications:
-
Kirby P. Hobbs, Amanda D. French, Kali M. Melby, Eric J. Bylaska, Khadouja Harouaka, Richard M Cox, Isaac J. Arnquist, and Chelsie L. Beck Analytical Chemistry 2024 96 (15), 5807-5814 DOI: 10.1021/acs.analchem.3c04774
-
Amanda D. French, Kali M. Melby, Kirby Hobbs, Richard M. Cox, Greg Eiden, Eric W. Hoppe, Isaac J. Arnquist, Khadouja Harouaka "The importance of ion kinetic energy for interference removal in ICP-MS/MS".Talanta,2024,125799, ISSN 0039-9140, https://doi.org/10.1016/j.talanta.2024.125799.
SimELIT: A Comprehensive Physics Based Ion Trajectory Simulation Software
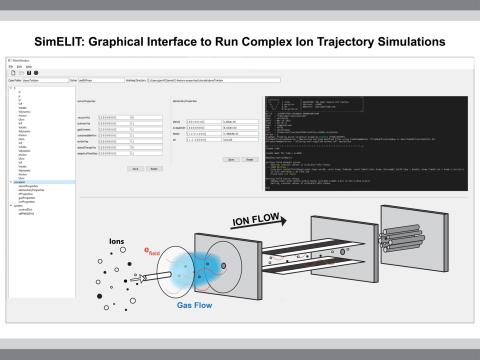
PI: Sandilya Garimella
Science Objectives
-
SimELIT: Simulator for Eulerian and Lagrangian Ion Trajectories – a novel modular, GUI based ion motion and trajectory simulation software
-
Accurate and comprehensive prediction of ion motion in mass spectrometry systems
-
Accurate ion trajectory calculation in multiphysics fields and multiple pressure scales can be leveraged for high performance computations of gas phase ion dynamics and chemistries inside mass spectrometry systems
Quantum Chemistry-Based Predictive Approaches for Ionization and Fragmentation
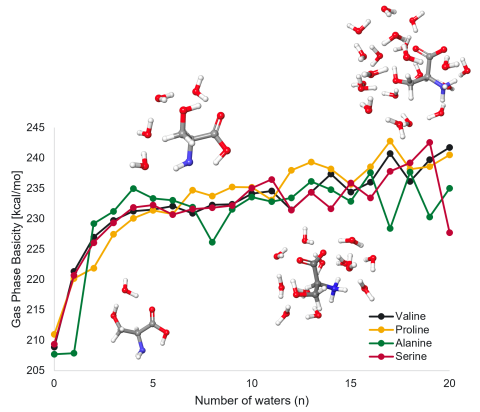
PI: Samantha Johnson
Science Objectives
- Robust and efficient computational workflow to understand and predict molecular ion generation during soft ionization methods
- Understanding of how droplet environment can alter the chemistry of molecules during mass spectrometry measurements
- Better understanding of how local environment affects structure and chemistry of molecules, with applicability beyond mass spectrometry (electrochemistry, gas-liquid interfaces, catalysis)
- Extending these simulations to enable quantitative relationships between matrix composition and observed spectra
A Joint Modeling/Experimental Approach to Characterize Ionization and Fragmentation of SOA Molecules with CIMS
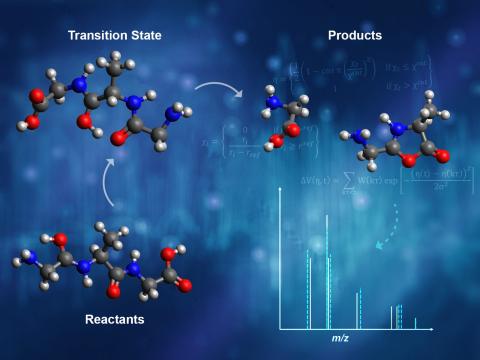
PI: Edoardo Aprà
Science Objectives
- Developed fast and accurate quantum mechanical simulation protocols to explore fragmentation pathways under soft ionization conditions
- Reference-free characterization of fragmentation pathways and product distributions to augment mass spectrometry spectral interpretation
- Extend the range of ionization and fragmentation processes relevant to mass spectrometry
- Include the effect of molecule-ion adducts and microsolvation conditions