Unlocking High-Efficiency Genome Engineering for Undomesticated Bacteria
SAGE is a high-efficiency genome integration strategy for bacteria that makes the stable introduction of new traits simple for newly discovered microbes
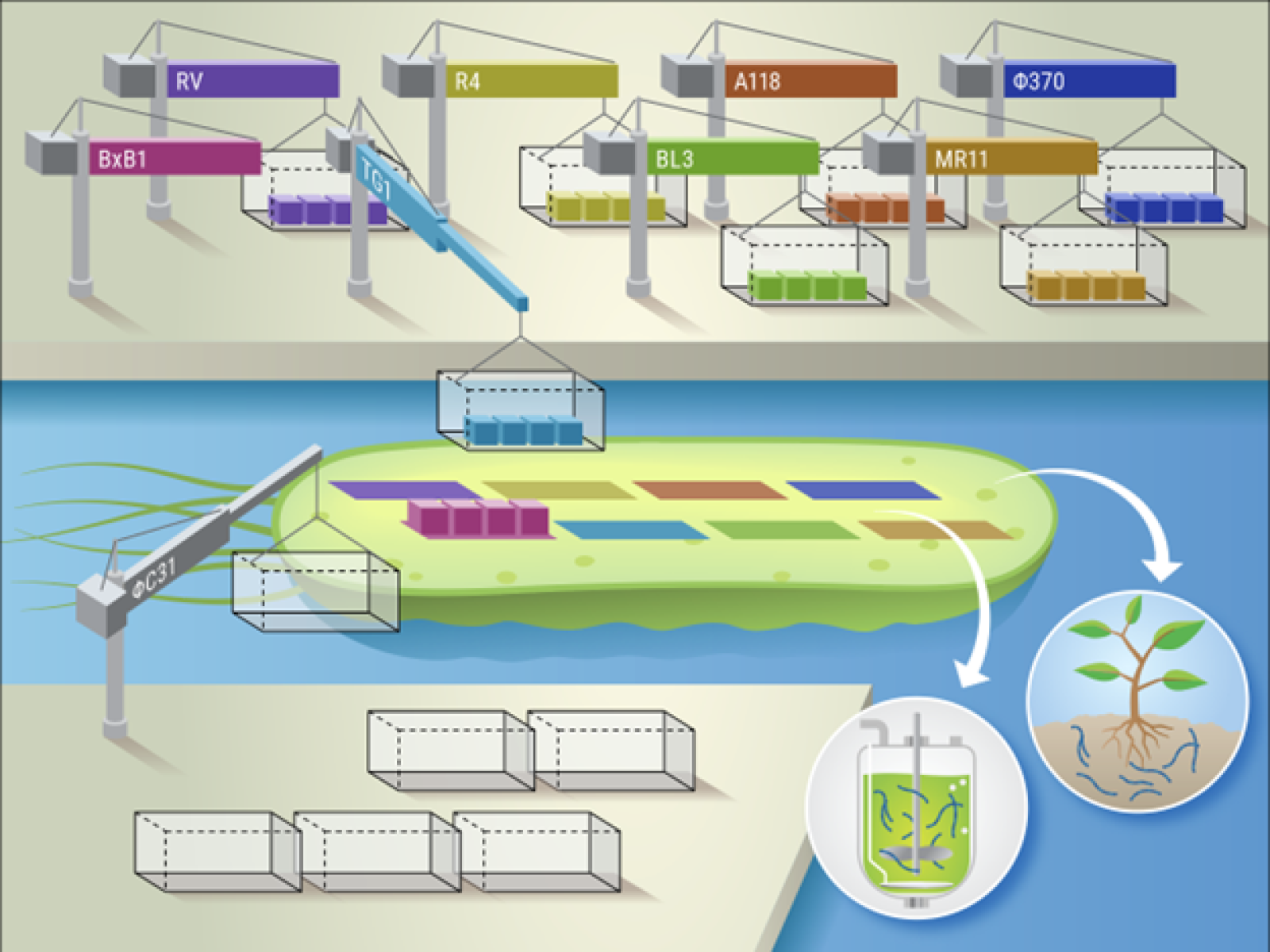
SAGE enables high-efficiency, multi-site integration of genetic cargo with recycled genetic containers. The efficient and stable integration of DNA circuits and metabolic pathways will advance the engineering of microorganisms for crop support and bioproduction.
(Illustration courtesy of Nathan Johnson | Pacific Northwest National Laboratory)
The Science
Introducing new genes to a microbe to produce novel functions is the essence of synthetic biology. Many genetic modification strategies lead to unstable expression outside of the laboratory and just do not work for most microbes that are brought into lab from the environment. Serine recombinase-Assisted Genome Engineering (SAGE) borrows components from viruses that attack bacteria to make it much easier to stably insert genes into bacterial chromosomes. SAGE genome integration components can be recycled to deliver multiple pieces of genetic cargo.
The Impact
Harnessing the potential of novel microbial isolates for biotechnology is significantly hampered by the lack of sophisticated genetic manipulation tools. A major factor in whether synthetic biology is successful is the stable transfer of engineered DNA into a microbial host cell. Most traditional genetic tools, such as small plasmid DNAs, are not stably maintained by cells, especially outside of laboratory environments. As a universal, efficient genetic manipulation system for bacteria, SAGE will accelerate the general goals of synthetic biology research. This includes engineering stable genetic programs to convert plant biomass into fuels, chemicals, and materials in fermentation processes, and engineering microbes that promote plant growth and human health.
Summary
Serine recombinase-Assisted Genome engineering (SAGE) provides a high-throughput genetic engineering toolkit for bacteria in any application space. By using up to 10 site-specific recombination pairs, genetic cargo can be stably installed on the chromosome. Multiple cycles of integration are possible by using different recombinases coupled with the recycling of selection and replication markers using a single excision recombinase. Together, the high-efficiency and multi-cycle integrations enable the installation of large genetic programs.
SAGE was demonstrated in five taxonomically diverse bacterial hosts that have attractive traits related to plant rhizosphere colonization and plant growth production, hydrogen production, bioremediation of hydrocarbons, and industrial bioproduction. In these hosts, SAGE provided transformation efficiency equivalent to or better than plasmid transformation for each host. Further, the integration of a library of taxonomically diverse bacterial promoter elements via SAGE revealed promoters with consistent behaviors across multiple environmental growth conditions.
PNNL Contact
Robert Egbert, Pacific Northwest National Laboratory, Robert.Egbert@pnnl.gov
Funding
This work was supported in part by the U.S. Department of Energy (DOE), Office of Biological and Environmental Research (BER), as part of BER’s Genomic Science Program (GSP) and is a contribution of the Pacific Northwest National Laboratory (PNNL) Secure Biosystems Design Science Focus Area "Persistence Control of Engineered Functions in Complex Soil Microbiomes". Additional support was provided by the DARPA Synergistic Discovery & Design program. PNNL is a multi-program national laboratory operated by Battelle for the DOE. This work was authored in part by Oak Ridge National Laboratory, which is managed by UT-Battelle, LLC, for the DOE. Funding was provided in part by the U.S. Department of Energy, Office of Energy Efficiency and Renewable Energy Bioenergy Technologies Office (BETO) to the Agile BioFoundry. This work was in part supported by the Center for Bioenergy Innovation, U.S. DOE Bioenergy Research Center, supported by the Office of Biological and Environmental Research in the DOE Office of Science.
Related Links
Published: April 26, 2023
Elmore, J. R., et al. (2023). "High-throughput genetic engineering of non-model and undomesticated bacteria via iterative site-specific genome integration." Sci Adv 9(10): eade1285. [https://doi.org/10.1126/sciadv.ade1285]