Peptide Fingerprinting Predicts Function using Protein Annotation Tool
Snekmer allows rapid prototyping to better understand protein function in microbes
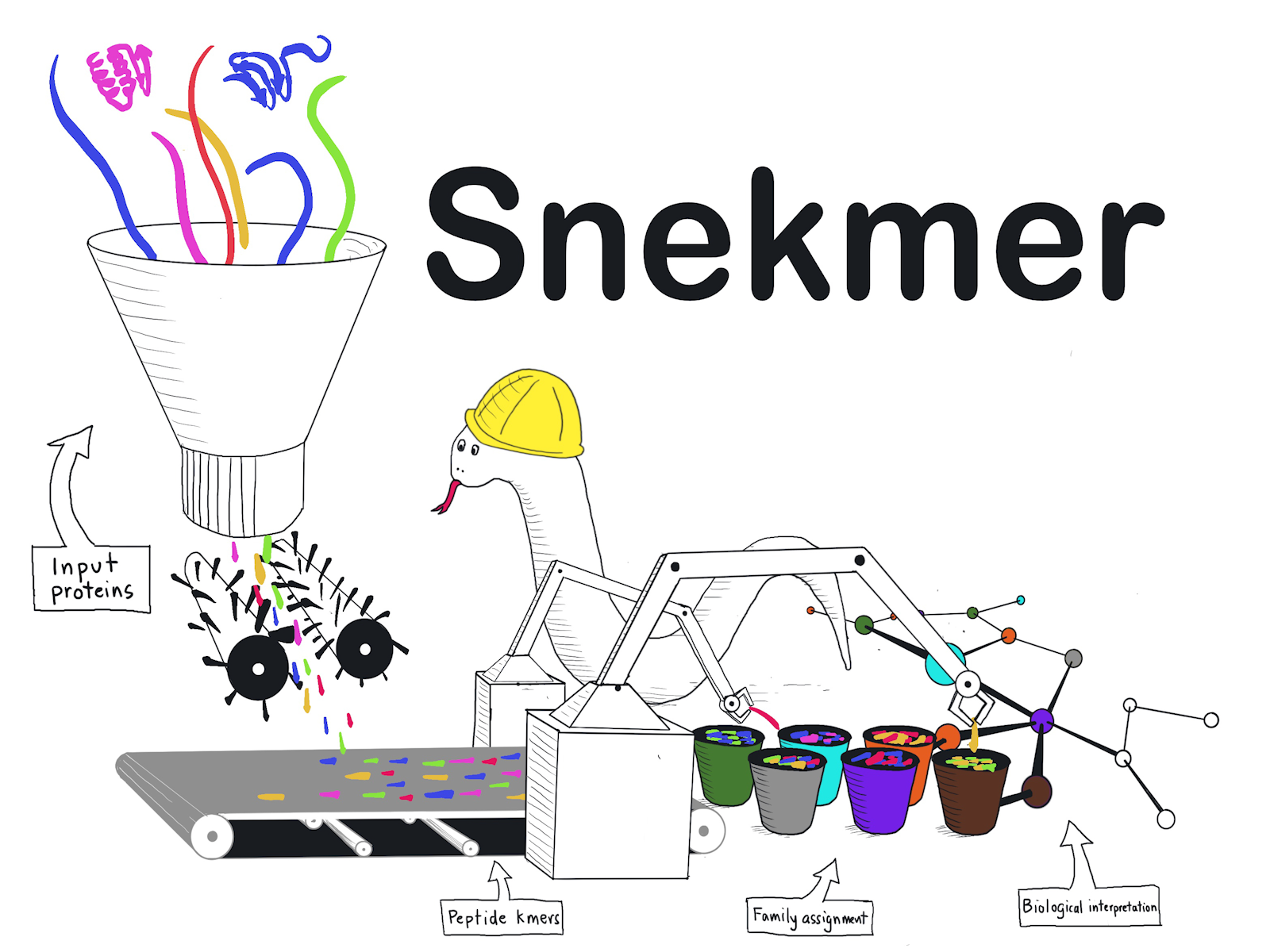
Snekmer is an application for building and searching protein family models and novel sequence clusters.
(Illustration by Jason McDermott | Pacific Northwest National Laboratory)
The Science
Microbes drive key aspects of global elemental cycles, promote plant growth, and affect the development of diseases. These roles are essential in every ecosystem. Research constantly expands the database of microbial DNA sequences but does not provide all the biological information about proteins. Our ability to engineer novel phenotypes to promote sustainable practices in bioenergy cropping and microbial bioconversion is limited by our understanding of molecular function.
The function of a protein can be inferred by comparing the sequence of a protein against reference databases of already characterized proteins. However, these comparisons are difficult and not scalable for massive databases. To address this challenge, we have applied machine learning to the deployment of models that predict protein function. Utilizing reduced-alphabet chemical feature representations of proteins, the program Snekmer allows rapid model prototyping and is available as a DOE KBase application.
The Impact
Collecting information and describing the biological molecules of protein ”dark matter” will facilitate a range of applications in applied science. Snekmer can be easily deployed in high-performance computing environments and is incorporated into the DOE KBase framework as a new application that will allow users to annotate their genome and metagenome sequences. Better climate impact modeling is possible, crop health and bioproduction are improved, and our understanding of evolutionary patterns and microbiome structure and function is expanded. This understanding of protein families is not limited to microbial systems.
Summary
The inability of current methods to predict function for 30–50 percent of bacterial protein sequences is a significant barrier to better understanding complex systems such as soil microbiomes. Most protocols rely on pair-wise alignments, which are becoming computationally intractable and more challenging to interpret as databases expand. For alignment-based models of protein families, the sensitivity and accuracy depend on the initial training sets, which risk obsolescence as additional sequence diversity is discovered. Many bacterial proteins have either no functional assignment or are only assigned a general function based solely on taxonomic understanding.
We have developed Snekmer, a software tool leveraging redundancy of amino acid residue properties to reduce sequence space and using short protein sequence (kmer) features for machine learning to generate protein family models. Snekmer users can recode protein sequences into reduced alphabet kmer vectors and perform the construction of supervised classification models trained on input protein families or protein functional classification based on Snekmer models.
PNNL Contact
Jason McDermott, Pacific Northwest National Laboratory, Jason.McDermott@pnnl.gov
Funding
This research was supported by the Department of Energy’s Biological and Environmental Research (BER) program and is a contribution of the Scientific Focus Area “Persistence Control of Engineered Functions in Complex Soil Microbiomes.”
Additional support was provided by the National Science Foundation (NSF) and the Defense Threat Reduction Agency (DTRA).
Published: March 10, 2023
Christine H Chang, William C Nelson, Abby Jerger, Aaron T Wright, Robert G Egbert, Jason E McDermott, Snekmer: a scalable pipeline for protein sequence fingerprinting based on amino acid recoding, Bioinformatics Advances, Volume 3, Issue 1, 2023, vbad005, doi.org/10.1093/bioadv/vbad005