LipidOz: New Software Enables Identification of Lipid Double Bond Locations
A streamlined informatics workflow that automatically processes complex data and provides the locations of double bonds in lipids
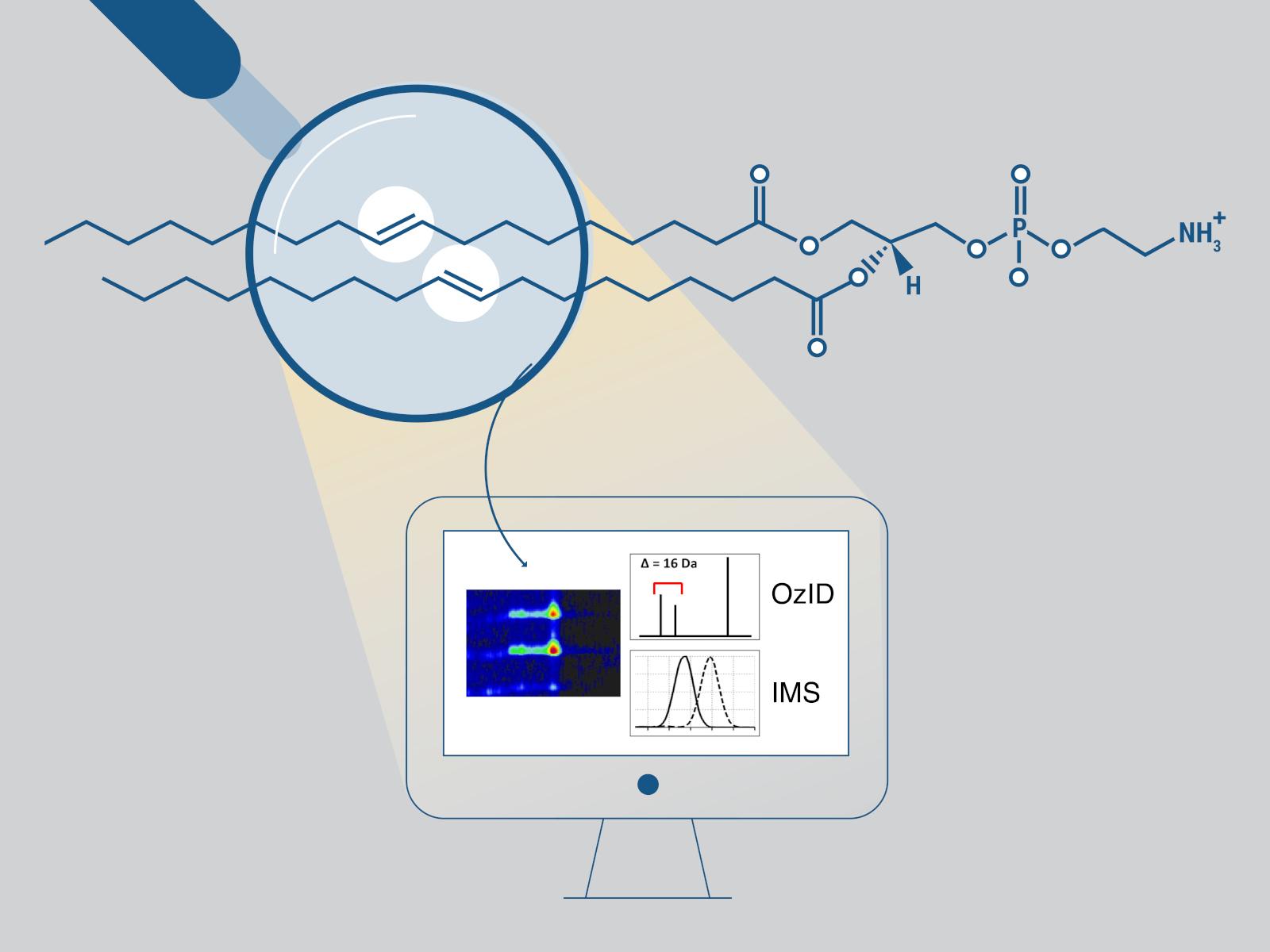
The LipidOz software helps researchers process the complicated data obtained from their instruments and assigns the locations of double bonds in lipids. Repetitive and difficult analyses can be automated with LipidOz.
(Image by Stephanie A. King and Nathan Johnson | Pacific Northwest National Laboratory)
The Science
Lipids are a class of biomolecules that play an important role in many cellular processes. Analyses that seek to characterize all lipids in a sample—called lipidomics—are crucial to studying complex biological systems. An important challenge in lipidomics is connecting the variety of structures of lipids with their biological functions. The positions of the double bonds within fatty acid chains is particularly important. This is because they can affect the physical properties of cellular membranes and modulate cell signaling pathways. This information is not routinely measured in lipidomics studies because it requires a complicated experimental setup that produces complex data. Thus, scientists at Pacific Northwest National Laboratory (PNNL) developed a streamlined workflow to determine the positions of double bonds. This workflow uses both automation and machine learning approaches.
The Impact
The ability to connect the structural characteristics of lipids to biology is often limited by the amount of structural information provided by analytical methods. The location of a double bond in a lipid is difficult to measure and requires complicated analytical approaches and specialized equipment. Even with access to this equipment, the available software lacks the ability to analyze the complex data. This severely limits the scope of studies that can be undertaken. LipidOz streamlines the data analysis to determine the positions of double bonds. By addressing this key part of the analysis of lipids, LipidOz offers researchers a more efficient and accurate method for lipid characterization.
Summary
Lipids play essential roles in many biological processes and disease pathology. However, the unambiguous identification of lipids is complicated by the presence of molecular parts that have the same chemical formula but different physical configurations. Specifically, the differences in these molecular parts include the fatty acyl chain length, stereospecifically numbered (sn) position, and position/stereochemistry of double bonds. Conventional analyses can determine the fatty acyl chain lengths, the number of double bonds, and—in some cases—the sn position but not the positions of carbon-carbon double bonds. The positions of these double bonds can be determined with greater confidence using a gas-phase oxidation reaction called ozone-induced dissociation (OzID), which produces characteristic fragments. However, the analysis of the data obtained from this reaction is complex and repetitive, and there is lack of software tool support. The open-source Python tool, LipidOz, developed in this work automatically determines and assigns the double bond positions of lipids using a combination of traditional automation and deep learning approaches. New research demonstrates this ability for standard lipid mixtures and complex lipid extracts, enabling practical application of OzID for future lipidomics studies.
Contact
Xueyun Zheng, Pacific Northwest National Laboratory, xueyun.zheng@pnnl.gov
Funding
This work was supported by the PNNL Laboratory Directed Research and Development program and is a contribution of the m/q Initiative. The establishment of the OzID-IMS capability was supported by the Laboratory Directed Research and Development Program at PNNL. Portions of this research were performed at EMSL, the Environmental Molecular Sciences Laboratory, a Department of Energy Office of Science user facility at PNNL which is sponsored by the Biological and Environmental Research program.
Published: January 16, 2024
Ross D. H., J.-Y. Lee, A. Bilbao, D. J. Orton, J. G. Eder, M. C. Burnet, B. L. Deatherage Kaiser, J. E. Kyle, and X. Zheng. 2023. “LipidOz enables automated elucidation of lipid carbon–carbon double bond positions from ozone-induced dissociation mass spectrometry data.” Communications Chemistry 6, 74. https://doi.org/10.1038/s42004-023-00867-9