Environmental
Molecular
Sciences Division
Environmental
Molecular
Sciences Division
The Environmental Molecular Sciences Division (EMSD) and the Environmental Molecular Sciences Laboratory (EMSL), the user facility that PNNL manages, support the Department of Energy (DOE) Office of Science's Biological and Environmental Research (BER) program mission to achieve fundamental understanding and prediction of complex biological, Earth, and environmental systems for energy and infrastructure security.
EMSD and EMSL are working toward three large-scale strategic science objectives.
- Digital Phenome Platform (DigiPhen): Engineering biology from biomolecules to organisms. This project is focused on creating methods for predictive modeling of protein function, pathway identification, and phenotype.
- Molecular Observation Network (MONet): Modeling from elements to ecosystems. This project creates a national molecular-level ecosystem/watershed/coastal monitoring network.
- Modeling and Data Sciences Capability (MDS): Visualizing all biological and environmental data. This project focuses on developing a BER-dedicated capability that provides mid-range high-performance computing for data-model integration and multiscale modeling.
The foundation for these long-term objectives are EMSD/EMSL's areas of expertise.
Biogeochemical Transformations | |
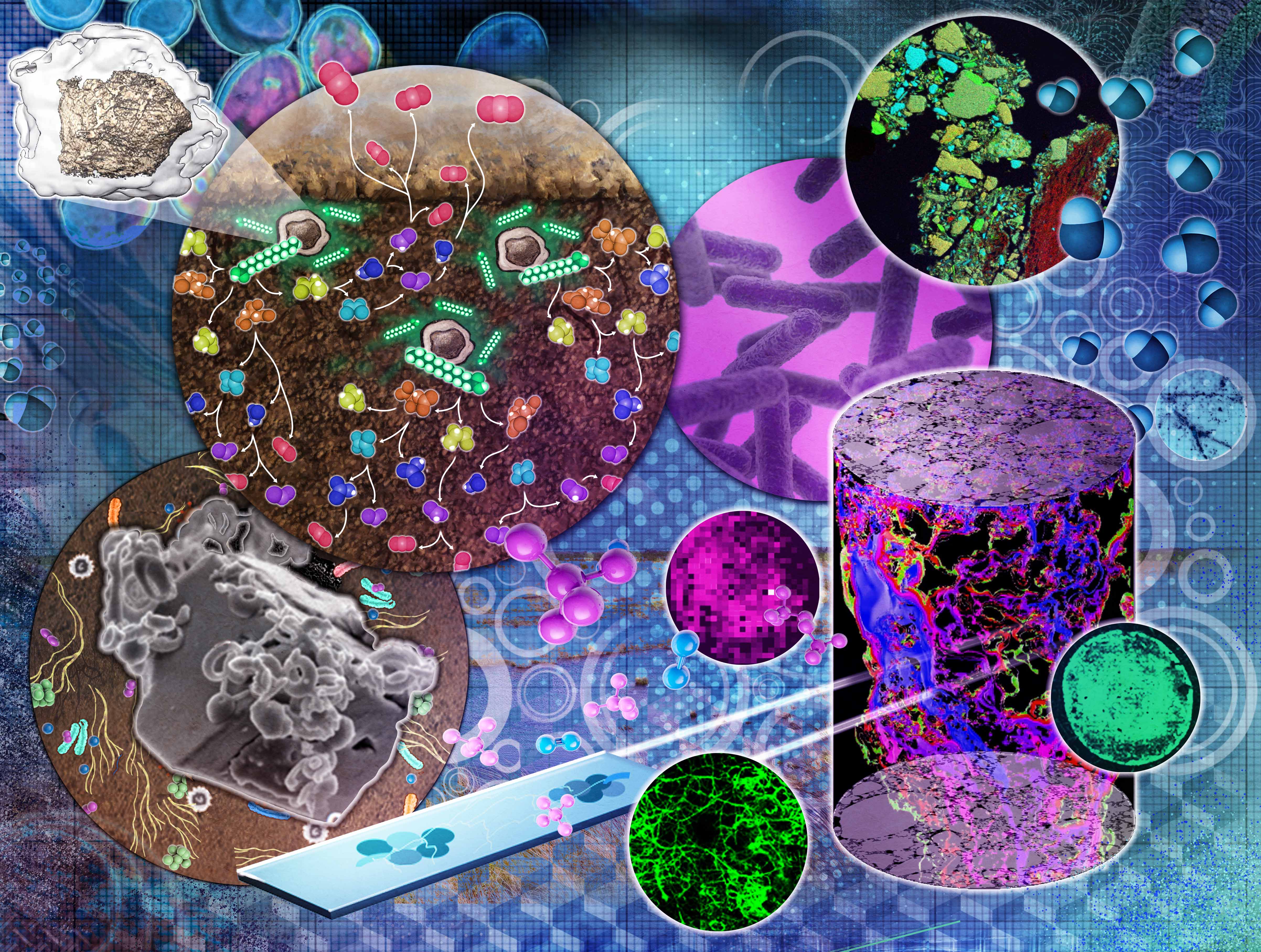 | The interplay of geology, chemistry, and biology among Earth systems is critically important for keeping Earth’s air and water clean and plants healthy. Our biogeochemical transformations expertise crosses scientific boundaries to investigate how nutrients, contaminants, aerosols, and other chemical compounds move and change in the environment. With new information on how these exchanges occur, we can develop and improve predictive models to anticipate and manage their effects on both human and natural systems. The Biogeochemical Transformations area of expertise helps researchers answer fundamental questions about chemical, physical, hydrologic, microbial, and atmospheric interactions that affect the transformation and mobility of critical nutrients, contaminants, aerosols, particles, and compounds within the environment. We use the latest platforms and approaches to gain new insights into the physiochemical effects of nutrient cycling in the environment, the interactions between metabolic pathways, and the dynamics of microbial communities. From molecular structures and activity, to cellular and community-scale processes, we seek to improve scientific understanding of microbial metabolism and nutrient cycling in the environment. |
Terrestrial-Atmospheric Processes | |
 | More than 28 percent of the Earth's surface is made of terrestrial ecosystems, or land-massed surfaces such as rainforests, deserts, and tundra. These ecosystems represent significant sources of atmospheric aerosols and chemical species, including pollen, soil minerals from rainfall, or volatile organic compounds from plants. Aerosols are important to controlling Earth's climate and are integral to hydrological and biogeochemical cycles. The Terrestrial-Atmospheric Processes area of expertise is dedicated to the study of terrestrial and atmospheric processes. Through this research area, scientists are addressing how aerosols and gases are emitted from plants and soil into the atmosphere. Researchers are examining how aerosols contribute to warm and cold cloud formation by behaving as cloud condensation nuclei or ice nucleating particles and how aerosols are deposited in terrestrial ecosystems. This will help scientists to develop experimental and observational data to support climate and Earth system models. |
Rhizosphere Function | |
 | The rhizosphere—the soil region around a plant's root—plays a vital role in biological, physical, and chemical processes that affect the integrated Earth system. Plant roots in the rhizosphere influence plant resilience, the formation of soil organic matter, and soil carbon sequestration. Scientists with the Rhizosphere Function area of expertise study the molecular mechanisms of interactions between roots, the soil, and microbes. Researchers also focus on how root-controlled processes affect biogeochemical nutrient cycling, the function and structure of microbial communities, and belowground carbon flux. Through research, scientists can improve understanding around the role of belowground processes in terrestrial carbon, relevant biogeochemical cycles, and coupled feedbacks to Earth and environmental systems. |
Structural Biology | |
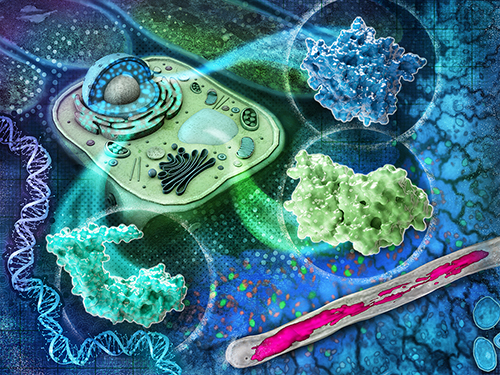 | All living systems rely on the tiniest of cellular functions to maintain health and vigor. When these functions break from disease or other stresses, the whole organism suffers. Structural biology allows us to study the relationships between cellular proteins, how they communicate, and their functions. With a better understanding of these cellular roles, we can look for ways to improve crops for food, biofuels, and bioproducts in a resource-constrained future. The Structural Biology area of expertise focuses on examining the assembly, structure, and function of proteins and protein complexes at the nanoscale, down to Ångströms, across space and time. With high-resolution images, we can see details in the molecular organization of a biosystem in three-dimensions. We can also map the chemical processes involved in micronutrient exchange both within and between cells and their environment, revealing links between proteins' structural and biochemical dynamics, protein complexes, and other biomolecules. These views help us understand how changes in morphology and composition affect biological systems. |
Biomolecular Pathways | |
 | Essential biological molecules—like proteins, lipids, RNA, and metabolites—create the language of gene expression and energy exchange that leads to healthy cells, organisms, and living systems. The Biomolecular Pathways area of expertise connects these biomolecules to their communication signals, biological roles, and energy functions to better explain and understand the trillions of small interactions that make up the world as we know it. This essential knowledge provides an enhanced understanding of cellular communication to improve resource use and create a more resilient environment. The collective biomolecules in plants, fungi, and microbes determine an organism’s structure, function, and dynamics. We have developed integrated capabilities to quantify the functional components of complex systems and probe biological molecules with unknown functions through increasing the rate, dynamic range, and resolution by which we analyze proteins and metabolites. Our accelerated throughput—with integrated transcriptomics, proteomics, and metabolomics—provides ultrasensitive measurements that explain the molecular mechanisms behind biological processes. With this capability, our research community explores biological complexity and diversity in the quest for more efficient biofuel production and robust environmental nutrient cycling predictions. |
Cell Signaling and Communication | |
 | Billions of cells, each with a one-of-a-kind role and behavior, make up living systems. Our expertise in systems biology, functional genomics, and biochemistry, enhanced by the state-of-the-art capabilities in high-resolution imaging and single-cell multi-omic measurements helps untangle the mechanisms by which cells communicate and exchange chemical signals. Quantitative studies of how individual cells grow and interact with their neighbors or hosts provide a critical understanding of molecular-level events underpinning complex biological systems' behaviors and their responses to the changing environment. The Cell Signaling and Communication area of expertise allows us to localize and isolate individual cells from complex biological samples for further structural and functional analyses. Researchers can then observe the live, intact cells and measure their unique responses over time to capture minute differences in cellular functions and outputs. Our technical capabilities enable labeling and characterization of subcellular structures and molecules in real time to quantify how, when, and where organisms exchange molecular signals and nutrients to foster interactions. As we begin to better understand and differentiate single-cell behavior, we are poised to predict functional outputs of interconnected biological systems as a function of their environment, thus enabling an informed design of organisms and communities with highly improved environmental and industrial properties for biosecurity and health applications. |
Data Transformations | |
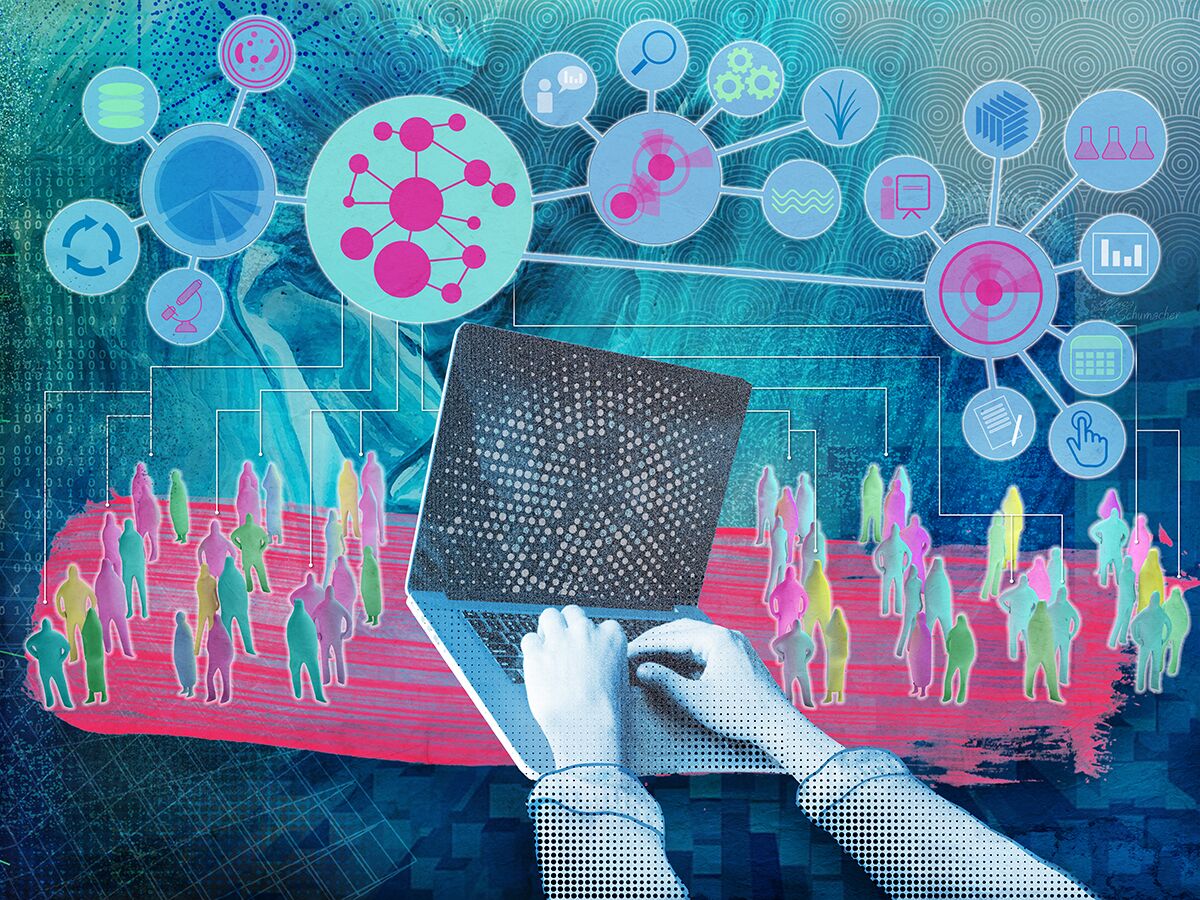 | Accessing and transforming data points into usable sets of information that showcase a process, structure, or system is challenging and complex. Biological and environmental researchers work with computational scientists in the Data Transformations area of expertise to streamline experimental setup in the computational space, identify ideal computational workflows, and transform the data into usable models and visualization tools. This collaborative approach enhances efficiency and effectiveness of scientific investigations, pushing the boundaries of knowledge and innovation. |
Systems Modeling | |
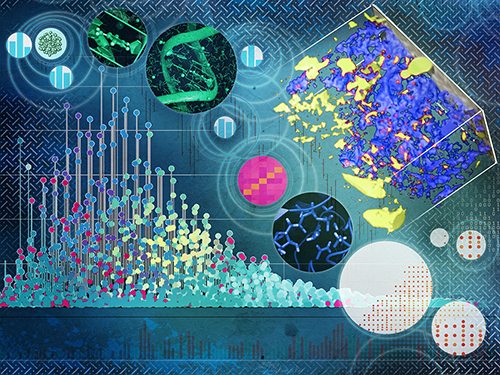 | Data is the foundation for advancing scientific discovery. Improved methods for producing, storing, and analyzing it are essential for expanding our knowledge of the world. With over 25 years of computing expertise, we're a leader in high-performance computing, software creation, and modeling development. Our primary science mission focuses on advancing the prediction and control of biological and environmental systems by developing advanced data analysis, data integration, multiscale modeling, and simulation approaches. At the heart of our Systems Modeling area of expertise is the model-experiment (ModEx) paradigm for biological and environmental systems. We are at the forefront of building computational models of protein structure and function, metabolic modeling, and machine learning techniques to associate genotype with phenotype, understand nutrient flux control processes, and create predictive biodesign and biofuel/bioproduct production approaches. Moreover, we track the flow of materials like carbon, nutrients, and contaminants in the environment to examine how biological and hydrobiogeological processes impact ecosystem function. |
Other Research
Scientists at EMSD also support non-BER-funded research projects that use EMSL's capabilities. Our scientists are leading or have led projects funded by:
- the National Science Foundation, including the Center for Sustainable Nanotechnology
- the National Institutes of Health, including the Pacific Northwest Center for Cryo-EM
- DOE’s Office of Energy Efficiency and Renewable Energy, including Next Generation Cathodes, Battery Materials Research, and Advanced Manufacturing Office
- NASA