A New Way to Identify and Control Specific Proteoforms
Scientists screen for nanobodies that recognize wild type and mutant functional proteins to develop a framework to disrupt protein interactions that can cause disease
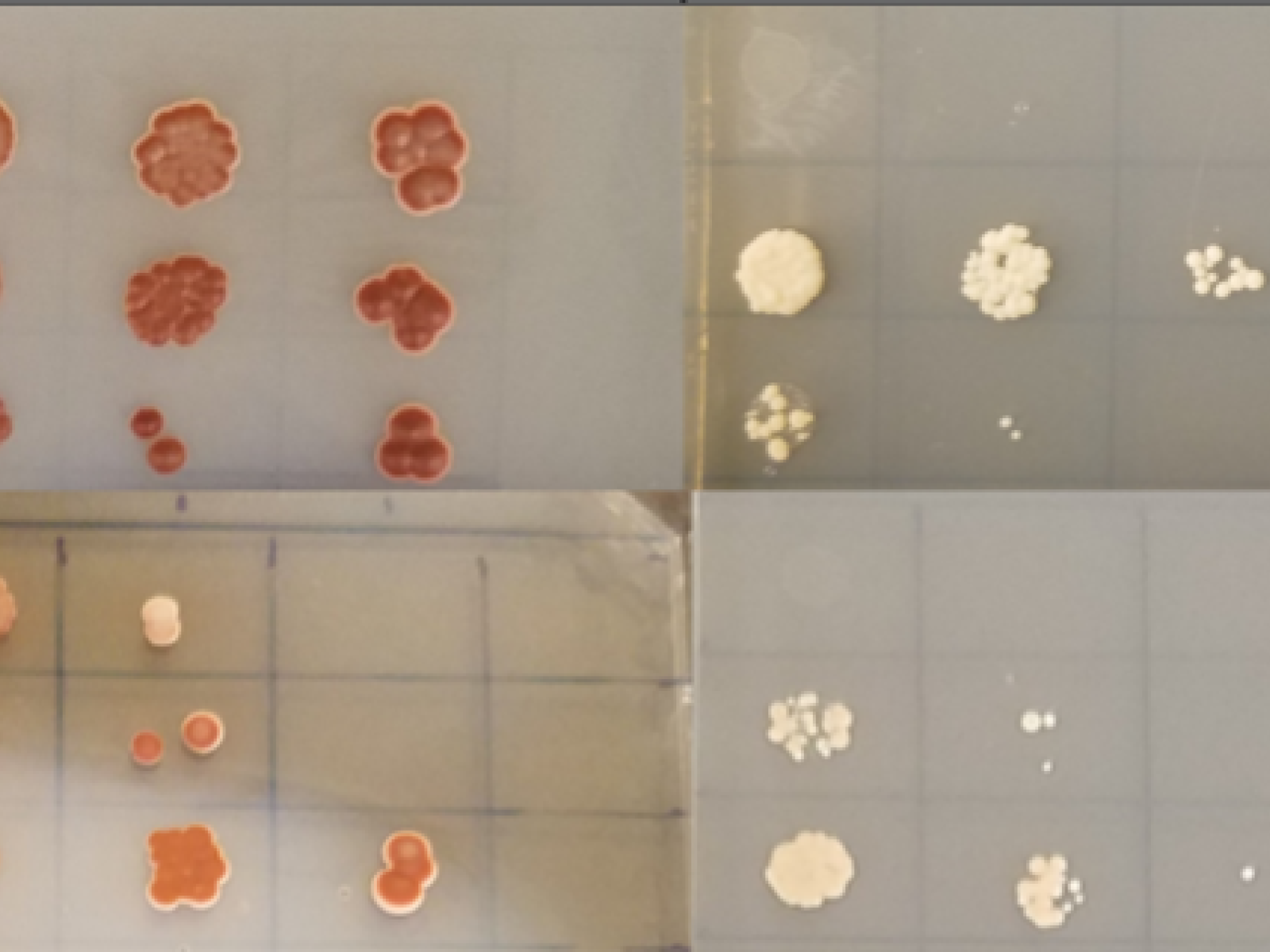
Confirming target protein and nanobody specificity: A yeast-2-hybrid experiment shows yeast growing on two selective media plates after mating. Growth of the yeast on the right requires expression and close interaction of the nanobody and protein target within the yeast.
(Image: Pacific Northwest National Laboratory)
The Science
Proteins are the work horses of the cell and mediate a wide variety of functions. A single protein can take many forms or produce variations known as proteoforms. Scientists have sought to identify and understand each modified form of proteins but, until now, proteoforms were extremely difficult to detect and differentiate from one another. In this study, scientists used yeast containing a nanobody library to identify nanobodies that bind and isolate specific proteoforms—a type of screening platform—which could eventually help identify novel protein functions to combat disease.
The Impact
Proteoforms, or proteins with different function-mediating structures, can provide clues about changes within our bodies that affect our health but they have been difficult to detect and analyze because of their complexity. Now, scientists have developed a new way to distinguish proteoforms from one another: training synthetic yeast nanobody libraries to bind to proteoforms of interest. This screening platform can be used in future studies for developing affinity reagents for purification of specific proteoforms. In addition, the results suggest that this platform can be used for the development of intrabodies to study specific proteoform functions inside cells where the protein normally functions.
Summary
Proteoforms expand genomic diversity and direct developmental processes. While high-resolution mass spectrometry has accelerated characterization of proteoforms, molecular techniques working to bind and disrupt the function of specific proteoforms have lagged.
In this study, scientists developed intrabodies capable of binding specific proteoforms. Once scientists isolated the individual proteoforms, they identified a framework for development of nanobodies and intrabodies that target them. A targeted approach of introducing antibodies into cells could disrupt proteoforms that lead to disease.
PNNL Contact
Amy Sims, Pacific Northwest National Laboratory, amy.sims@pnnl.gov
Funding
This project was funded by the Congressionally Directed Medical Research Program award CDMRPL-20-0-PR201356 and the PNNL Predictive Phenomics Initiative LDRD. PNNL is a multiprogram national laboratory operated for the U.S. Department of Energy (DOE) by Battelle Memorial Institute. This research used computational resources provided by Research Computing at the Pacific Northwest National Laboratory.
Published: September 29, 2023
B. Leonard, Danna, V., Gorham, L., Davison, M., Chrisler, W., Kim, D.M., and Gerbasi, V.R. 2023. “Shaping Nanobodies and Intrabodies against Proteoforms.” Analytical Chemistry 2023 95 (23), 8747-8751. DOI: 10.1021/acs.analchem.3c00958