Making a Protein “Multitool” for Assembly
Changing the environment of proteins directs their assembly into different types of larger structures
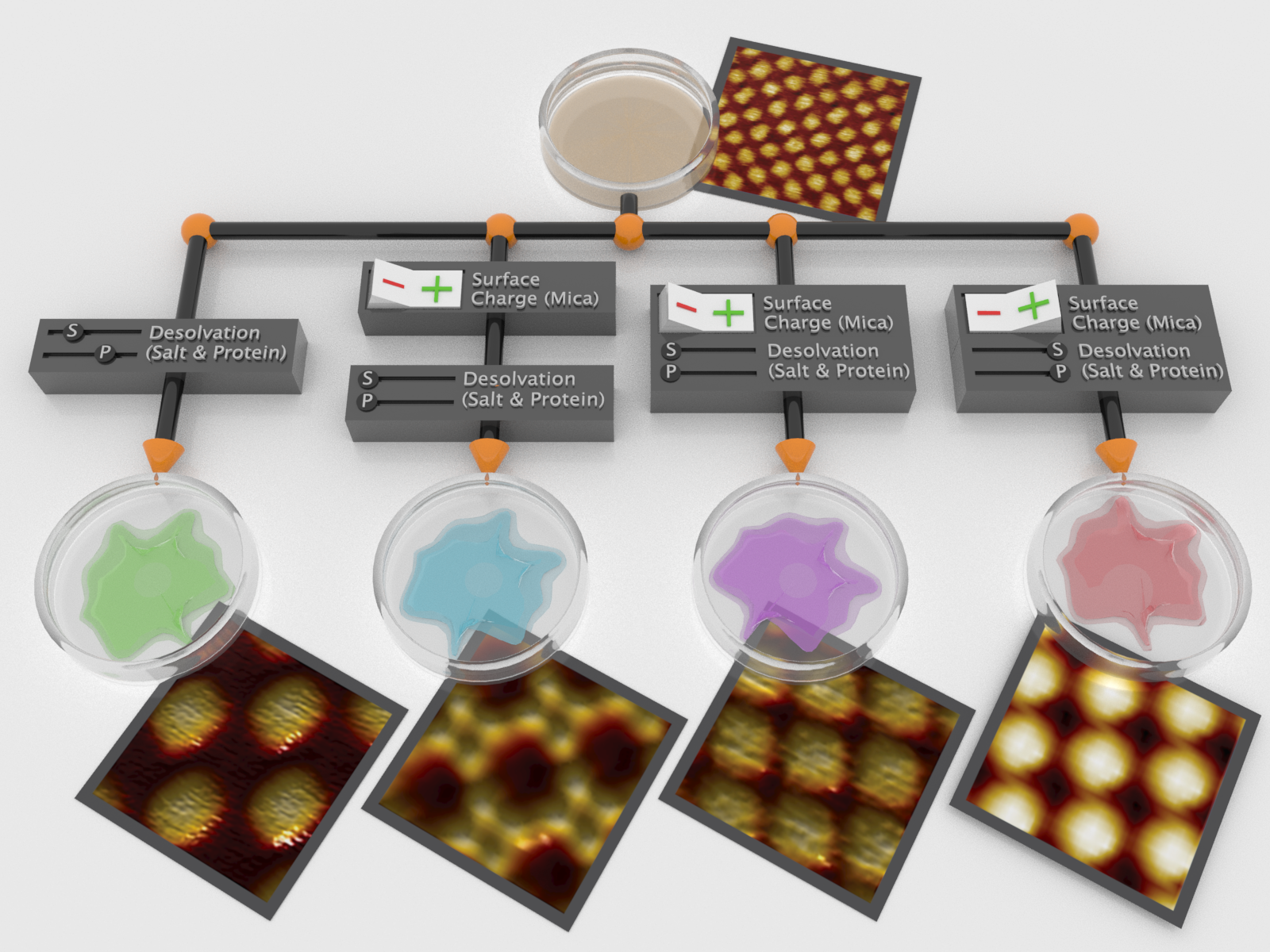
By using charged mineral surfaces and aqueous electrolytes to manipulate interactions over many length scales, a single type of protein is driven to self-assemble into multiple, diverse lattices with novel properties.
Image: Nathan Johnson | PNNL
The Science
Many materials are made from the same atoms or molecules but have very different bulk-scale properties. A common example of this is diamond vs graphite: both made of just carbon atoms, but the differences in bonding make the former extremely hard and the latter soft. This research set out to understand how proteins could similarly be used to form different materials with unique properties from a single building block. There has been much scientific effort aimed at understanding the design principles necessary for creating proteins purpose-built for technological applications. Many natural proteins can serve in multiple roles in different contexts, but until now engineered (“designed”) proteins typically only achieved a single type of structure or function. This research clearly demonstrated that it is indeed possible to create a protein “multitool” that can be arranged into new and useful ways depending on its environment. Scientists achieved synthetic control over protein self-assembly pathways by altering the effects of four kinds of interactions on the assembly of a single patchy protein. This allowed the realization of four distinct 2D crystal morphologies with similarly novel properties.
The Impact
This work demonstrates that knowledge from “patchy nanoparticle” self-assembly can be extended to proteins to produce new architectures with specific material properties. By balancing the strengths and length scales of different interactions, multiple novel assemblies can be created from a designed protein. The assembled crystals also exhibit electronically interesting behavior not found in the original protein. Their permanent electrical charge distributions combined with the porous structures of the assemblies makes these materials attractive for applications ranging from water purification systems to nanoscale electrical generators. They pave the way for work utilizing other designed proteins to access more structures with unique functionalities.
Summary
The self-assembly of molecular building blocks into higher-order structures is a powerful strategy for constructing new materials. The identical nature and atomically tunable interactions of proteins make them unique as nanoscale building blocks. This report shows the modular self-assembly of an engineered protein into four physicochemically distinct, precisely patterned 2D crystals via control of four classes of interactions (dipole-dipole, electrostatic, disulfide bonding, and desolvation interactions) spanning Ångström to tens-of-nanometer length scales. The resulting structures are related to the underlying free-energy landscape by combining in-situ atomic force microscopy observations of assembly with thermodynamic analyses of protein-protein and -surface interactions. The results demonstrate the rich phase behavior obtainable from a single, highly patchy protein when interactions acting over multiple length scales are exploited and predict unusual bulk-scale properties for protein-based materials that ensue from such control.
PNNL Contacts
James De Yoreo, Pacific Northwest National Laboratory, James.DeYoreo@pnnl.gov
Funding
This work was supported by Center for the Science of Synthesis Across Scales (CSSAS).
Published: September 25, 2020
Zhang S, R G Alberstein, J J De Yoreo, F Akif Tezcan. 2020. “Assembly of a patchy protein into variable 2D lattices via tunable multiscale interactions.” —Nature Communications, 11:3770. DOI: 10.1038/s41467-020-17562-1.