Cyanobacteria Rapidly Adapt to Environmental Perturbations Through Structural Remodeling of the Proteome
Structural proteomics reveal how cyanobacteria adapt to changing light
Understanding molecular modifications in red yeast could help scientists re-engineer the organism’s phenotype.
(Image by Stephanie King | Pacific Northwest National Laboratory)
The Science
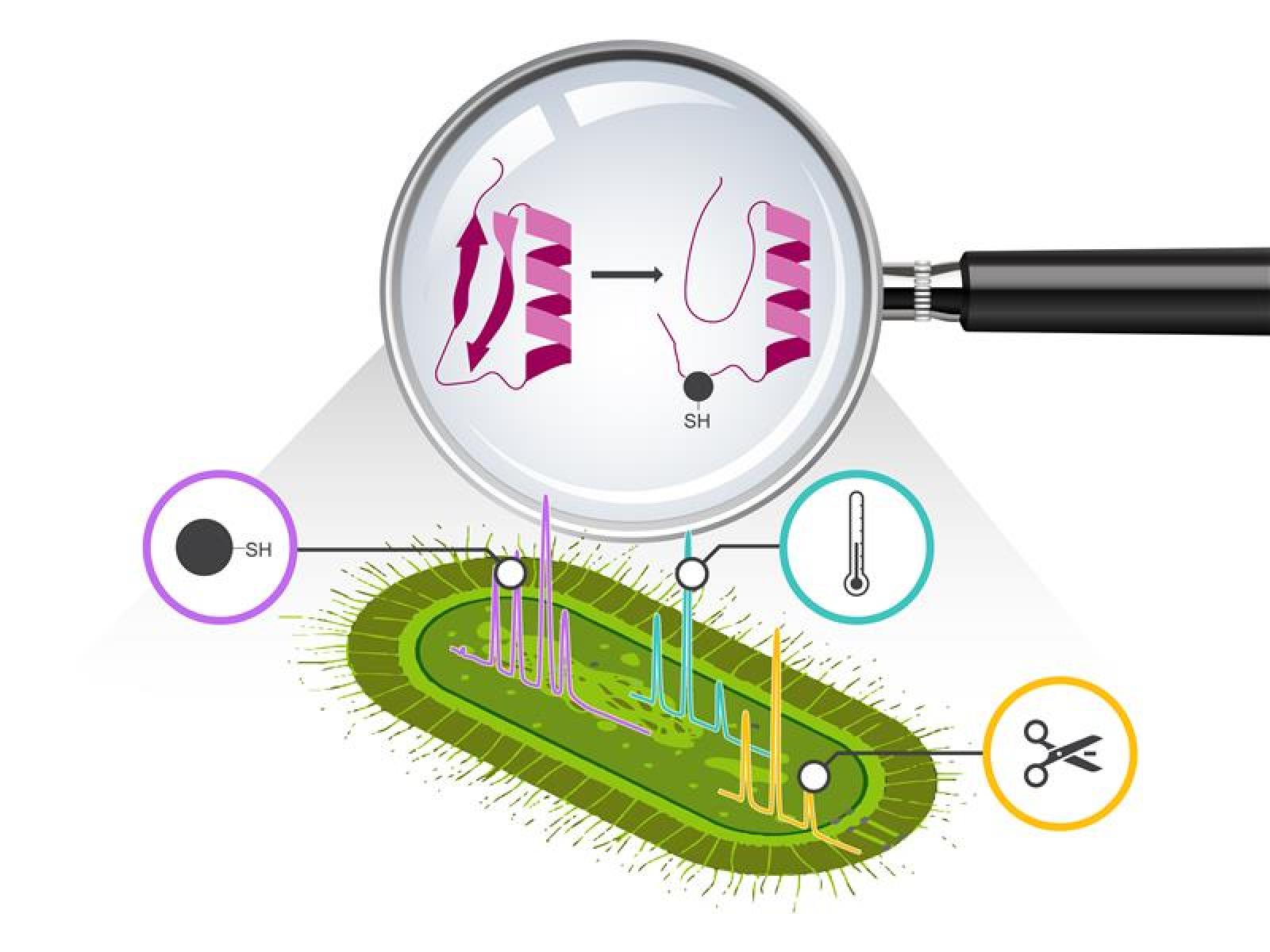
Understanding how organisms like cyanobacteria adapt so quickly to changes in their environment is a significant challenge. Traditional systems biology approaches using proteomics generally track the quantities of protein present but miss dynamic shifts in protein structure and activity that happen long before new proteins are made or old ones broken down.
This study, supported by the Predictive Phenomics Initiative, reveals that cyanobacteria rapidly adapt to changing light by extensively and quickly reshaping their existing proteins. These structural changes happen on a faster time scale than protein synthesis or degradation and directly affect key cellular processes such as photosynthesis, energy metabolism, and protein translation. By unveiling that fast, reversible protein structural changes drive much of the early adaptive response, this work provides a new view of how cyanobacteria reprogram their proteome to survive and function under fluctuating environmental conditions.
The Impact
This work addresses a central problem in biology and biotechnology: organisms respond quickly to changing conditions, but researchers have lacked a detailed, system‑wide view of the fast structural protein changes that make this possible. By showing that cyanobacteria rapidly remodel the structures, stability, and redox states of thousands of proteins well before protein levels change, this study provides one of the first comprehensive, proteome‑wide maps of how structural protein dynamics drive environmental adaptation in a photosynthetic microbe. Innovative in both scope and approach, the study combines four high‑throughput proteomics measurements in a single, coordinated experiment on the same biological system. Each method detects a largely distinct set of responsive proteins, highlighting that no single technology can adequately capture the complexity of the cellular response.
These findings open the door to building more accurate predictive models of how microbial cells reprogram themselves under biomanufacturing and environmentally relevant fluctuating conditions. In practical terms, this work enables the field to (1) better identify which proteins and pathways control rapid acclimation of photosynthesis and metabolism, (2) rationally engineer cyanobacteria and other microbes for more robust and efficient production of biofuels and bioproducts, and (3) apply similar multi‑modal proteomics strategies to other organisms, including plants and human cells, to understand fast stress and signaling responses at a systems level.
Summary
Cyanobacteria live in environments where light conditions can change from moment to moment, yet maintain efficient photosynthesis and growth. To understand how they do this, researchers studied how the model cyanobacterium Synechococcus elongatus PCC 7942 responds when shifted from low to high light. Traditional proteomics showed that only a modest number of proteins changed in abundance in the first 30 minutes. However, examining protein structure, stability, and redox state showed thousands of additional, rapid changes across the proteome.
These results demonstrate that cyanobacteria rely heavily on fast, reversible modifications of existing proteins missed by techniques that measure the slower changes in protein production or degradation. These structural proteomic changes quickly adjust key processes such as photosynthesis, energy metabolism, and translation. By combining multiple, complementary proteomics methods in a single experiment, researchers generated a detailed, system‑wide view of how the structural proteome is reprogrammed during early light acclimation. This provides a new framework for understanding and eventually predicting how photosynthetic microbes adapt to the fluctuating environments relevant to both natural ecosystems and biomanufacturing.
Contact
John Melchior, john.melchior@pnnl.gov, Pacific Northwest National Laboratory
Funding
The research described in this paper is part of the Predictive Phenomics Initiative at Pacific Northwest National Laboratory (PNNL) and conducted under the Laboratory Directed Research and Development Program. PNNL is a multiprogram national laboratory operated by Battelle for the U.S. Department of Energy under Contract No. DE-AC05-76RL01830.
Published: February 11, 2026
Sarkar, S. et al. 2025. “Rapid adaptation of cyanobacteria to environmental perturbations is achieved through structural remodeling of the proteome.” Mol Cell Proteomics, 101443. doi: 10.1016/j.mcpro.2025.101443