Accounting for Solvent Polarization Improves Conformational Predictions in a Peptoid Force Field
A new peptoid force field better predicts the cis/trans equilibrium of peptoids in multiple solvents
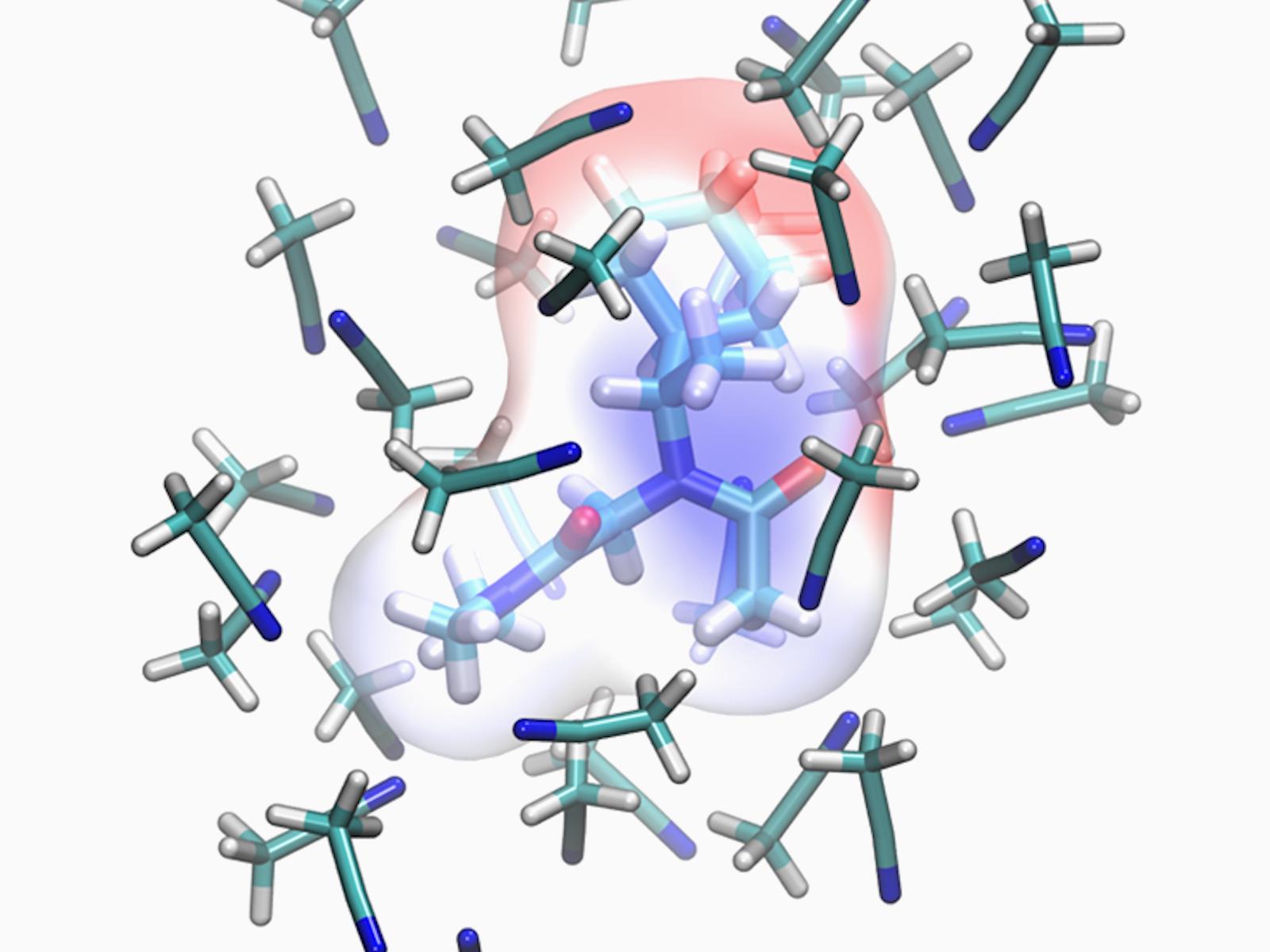
Electrostatic potential mapped on the van der Waals surface of the R57 peptoid monomer in acetonitrile, highlighting the solvent-dependent charge distribution used in STEPs-SOL.
(Image by Marcel Baer | Pacific Northwest National Laboratory)
The Science
Peptoids are a promising class of biomimetic molecules that are more stable and tunable than natural peptides. Designing larger materials from peptoids requires molecular models that capture peptoid conformational preferences. Researchers showed that accounting for solvent-dependent polarization is essential for accurate simulations of peptoid cis/trans equilibria. To incorporate this physics, the team developed a new solvent-adaptive extension of the Systematic and Extensible Force Field for Peptoids, named STEPs-SOL. By deriving solvent-polarized Restrained Electrostatic Potential (RESP) charges from representative solvated backbone and side-chain conformations, STEPs-SOL improves agreement with experimental cis/trans ratios across multiple solvents. It reduces errors by more than 30 percent compared to prior force fields that neglect solvent polarization.
The Impact
Accurately modeling peptoid conformational preferences is necessary for researchers to design biomimetic polymers with tailored functions. The newly developed STEPs-SOL force field enhances simulation accuracy, enabling a more reliable representation of peptoid structure across a wide range of solvent conditions. By replacing a reliance on the small set of experimentally determined equilibrium constants with a charge-derivation workflow based on solvated conformations, STEPs-SOL enables predictive simulations of sequence-defined peptoid materials. This advance opens the door to rational design of biomimetic polymers with tailored functions—whether targeting specific folding, assembly, or interaction properties—using highly automated, reproducible computational pipelines.
Summary
Peptoids can be used to create tunable, precisely defined hierarchical biomimetic materials. To design peptoid-based materials for specific applications more quickly and efficiently, researchers need to accurately model the structure and behavior of peptoids. STEPs-SOL represents a step change in peptoid modeling by explicitly incorporating solvent polarization into a fixed-charge force field. The workflow begins with exhaustive sampling of the twelve distinct minima in backbone dihedral space, together with systematic rotations of side-chain rotamers. The resulting structures are clustered to identify representative low-energy conformers. These conformers are then placed in explicit solvent boxes of water, methanol, chloroform, or acetonitrile and simulated to capture solvent effects. Electrostatic potentials from the solvated snapshots are fit with the IPolQ RESP protocol to obtain solvent-polarized charges. Compared to the original vacuum-based STEPs charges, STEPs-SOL reduces the mean absolute error in cis/trans free-energy predictions by more than 30 percent, delivering much closer agreement with experimental nuclear magnetic resonance spectroscopy data. STEPs-SOL can help enable truly predictive simulations of sequence-defined peptoid materials by accurately representing the material energies. This advance takes a step toward allowing researchers to leverage highly automated, reproducible computational pipelines to design peptoid-based materials with specifically targeted functionalities.
Contact
Marcel Baer, Pacific Northwest National Laboratory, marcel.baer@pnnl.gov
Funding
This work was supported by the Department of Energy, Office of Science Early Career Research Program and Basic Energy Sciences, Materials Sciences and Engineering Division, Biomolecular Materials Program (primary support FWP77876). C.J.M. and Y.W.E. also acknowledge FWP80124 for their efforts in implementing the workflow and generating data. Computing resources were provided by the National Energy Research Scientific Computing Center, an Office of Science User Facility at Lawrence Berkeley National Laboratory.
Published: September 12, 2025
Elhady, Y. W., B. S. Harris, C. J. Mundy, M. D. Baer. “STEPs-SOL, a Peptoid Force Field Parameterization to Include Solvent Effects,” J. Phys. Chem. B, 129, 5901 (2025). [DOI: 10.1021/acs.jpcb.5c02834]